![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
307 Cards in this Set
- Front
- Back
|
What are the basic steps of the scientific method? What happens at each step?
|
1. Observation – look at what’s going on
2. Hypothesis – an educated guess of the results 3. Check predictions – use hypotheses to test your predictions 4. Conclusion – record your results; what hypothesis was/was not rejected. |
|
|
We can divide organisms into what was historically recognized as the five major kingdoms. What are the names of these kingdoms, what are general characteristics of each kingdom? From Kingdom to Species, what are the different categories of classification?
|
1. Bacteria – prokaryotic cells; anaerobic/aerobic metabolisms; heterotrophic/autotrophic feeding; cell walls with peptidoglycans; susceptible to many antibiotics.
2. Archea – prokaryotic cells; anerobic/aerobic metabolisms; heterotrophic/autotrophic feeding; cell walls lack peptidoglycans; not susceptible to many antibiotics. 3. Eukarya-Protista – very diverse eukaryotic organisms that are not plants, fungi, or aninals; may single-celled species; mostly aerobic; autotrophic/heterotrophic. 4. Eukarya-Fungi – eukaryotic cells will walls containing chitin; aerobic/anerobic; mostly multicellular; hetertrophic; important recycling agents (sac and club fungi, molds, yeast). 5. Eukarya-Plantea – eukaryotic cells with walls containing cellulose; mostly aerobic; mostly autotrophic; multicellular; mostly immobile (mosses, ferns, flowering plants, pines). 6. Eukarya-Animalia – eukarytic cells without cell walls; aerobic; rarely anaerobic; mostly multicellular; heterotrophic; many possess nerve and muscle tissue (sponges, worms, mollusks, insects, vertebreas). 7. Domain > Kingdom > Phylum > Class > Order > Family > Genus > Species |
|
|
How can you differentiate among covalent, polar covalent and ionic bonds? Give two examples of two types of bonds that are weaker than covalent and ionic bonds.
|
A covalent bond is formed between two atoms that share electrons equally. A polar covalent bond is formed between two atoms that share electrons unequally so that one atom is relatively more negative than the other atom. An ionic bond is formed between two atoms when one atom completely pulls an electron from the second atom and the two atoms are held together because one is now negatively charged (containing an extra electron) and the other is positively charged (because it has lost an electron). Covalent bond - gaseous hydrogen, H2 or oxygen, O2. Polar covalent bond - H2O. Ionic - NaCl. In general, the number of covalent bonds an atom can form is equal to the number of unpaired electrons in has in its outer shell. Oxygen has an atomic number of 8 and therefore it has 6 electrons in its outer shell. It has room for two electrons in its outer shell and can form two covalent bonds. In the case of O2, each oxygen atom has two unpaired electrons and the electrons of one atom are paired with the other atom completing the outer shell of each. Note, this requires then a double bond between the two oxygen atoms. Carbon has 4 unpaired electrons and therefore it shares two electrons with each of the oxygen atoms in CO2. Thus there is a double bond between carbon and each oxygen.
|
|
|
What kind of bonds hold the atoms of a water molecule together? What kind of bonds hold water molecules together? How do these bonds between atoms produce the bonds between water molecules?
|
1. Hydrogen bonds hold the atoms of water molecules together.
2. Polar covalent bonds hold water molecules together. 3. Polar covalent bonds produce relatively positive and negative ends to the water molecule. |
|
|
How does a hydrogen ion differ from a hydrogen atom? Why are the terms hydrogen ion and proton sometimes used interchangeably? Define the terms acid, base and buffer.
|
1. Hydrogen ions (H+) differ from hydrogen atoms because hydrogen ions have a single proton charge of 1+.
2. The water molecule that loses a proton is now a hydroxide ion (OH-). 3. Acid – a substance that increases the hydrogen ion concentration of a solution. 4. Base – a substance that reduces the hydrogen ion concentration of a solution. 5. Buffer – substances that minimize the changes in H+ and OH- in a solution. |
|
|
What are the expected, molar concentrations of hydrogen ions (H+) and of hydroxyl ions (OH-) in pure water? What is the definition pH? What are the molar concentrations of hydrogen ions (H+) in solutions with pHs of 8, 7, and 2? What would be the [OH-] in these 3 solutions? At what pH do most biological systems function best?
|
1. The expected molar concentrations of H+ and OH- ions in pure water is 1x10-7 M.
2. pH – the negative of the log of [H+] >> pH=-log[H+] 3. for [H+] concentrations >> pH of 8 = 1x10-8M; pH of 7 = 1x10-7M; pH of 2 = 1x10-2M 4. Each should be 1 X 10-7 M. pH = -log [H+] . [H+] in the following solutions: pH 8= 1 X 10-8 M, pH 7 = 1 X 10-7 M, pH 2= 1 X 10-2 M. The [OH-] concentration in each of these solutions would be: 1 X 10-6 M, 1 X 10-7 M, and 1 X 10-12 M. Most biological systems operate best at near neutral pH. (pH=7). |
|
|
Briefly describe the structure and function of starch and glycogen. In what type of organisms are these molecules manufactured?
|
Starch and glycogen store energy for organisms. Starch is primarily found in plants and glycogen is found in animals.
|
|
|
Define the terms atomic number, atomic mass and atomic weight. What elements are represented by the chemical symbols C, H, O, N, S, P, Na, K, Cl, Ca, Fe, Mg Cu, B? What are isotopes and radioisotopes?
|
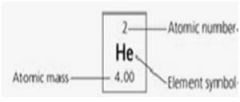
1. Atomic number – the number of protons in an atom.
2. Atomic mass – approximately the mass number of an atom in Daltons. 3. Atomic weight – the number of protons and neutrons in an atom. 4. Carbon, hydrogen, oxygen, nitrogen, sulfur, phosphorus, sodium, potassium, chlorine, calcium, magnesium, copper, boron. 5. Isotopes – Atoms with the same atomic number but different mass numbers. 6. Radioactive isotopes – unstable isotope that decays after giving off particles with energy. |
|
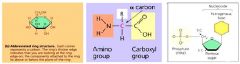
Compare figures 5.4b (glucose, a monosaccharide), 5.17 (amino acids) and 5.26b (a nucleotide). How can you distinguish them from one another? What kind of polymers does each of these monomers form? What are dehydration (condensation) and hydrolysis reactions and how do they relate to the formation of polymers from monosaccharides and amino acids? What is a glycosidic bond? What is a peptide bond? The molecular formula for glucose is C6H12O6. What would be the molecular weight of one glucose molecule? What would be the molecular weight of 10 glucose molecules if they were polymerized by dehydration reactions?
|
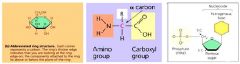
1. Glucose, a monosaccharide, generally have molecular formulas that are some multiple of the unit CH2O, the molecules has a carbonyl groups (>C=O) and multiple hydroxyl groups (-OH); amino acids possess both carboxyl and amino groups; nucleotides are made of three components a.) nitrogenous base b.) a sugar and c.) a phosphate group.
2. Glucose forms – most sugars found in body; amino acid forms – produce amino acid groups; nucleotide forms – the DNA and RNA. 3. Dehydration (condensation) reactions – polymers lengthened by addition of monomer which requires the loss of water molecules. 4. Hydrolysis – polymers shortened with release of a monomer and addition of a water molecule. 5. Glycosidic bond – a covalent bond formed between two monosaccharides by dehydration. 6. Peptide bond – the covalent bond between two amino acid units, formed by a dehydration reaction. 7. The molecular weight of a glucose molecules = 155.88g; the molecular weight of 10 glucose molecules = 1558.8g. |
|
|
Define diffusion, selectively permeable membrane and osmosis. What do the terms hypotonic and hypertonic mean? What is the direction of osmotic water movement, relative to these terms? Would you expect a healthy plant cell to be hypertonic, hypotonic or isotonic relative to its environment? Why? What about a typical animal cell? Why? What happens to plant or animal cells when they are placed in a hypotonic solution? A hypertonic solution?
|
1. Diffusion – passive transport of solute molecules from areas of high concentration to low concentration. Selectively permeable membrane – control transport into and out of the cell. Osmosis – diffusion of water across a selective permeable membrane.
2. Hypotonic = low concentration; Hypertonic = high concentration. 3. The direction of osmotic water is from a low (hypotonic) to high (hypertonic) solute concentration. 4. Plant cells would be hypertonic because water is moving into the cells. 5. Animal cells would be isotonic because there is no net movement of water. |
|
|
Define facilitated diffusion and active transport. What is an ATPase and how does it relate to active transport? What is an electrogenic pump?
|
1. Facilitated diffusion – passive transport of solutes across a membrane (down a concentration gradient) through proteins; Active Transport – transport of solutes across a membrane against a concentration gradient with the help of energy input and transport proteins.
2. ATPase is an enzyme that breaks down ATP; ATP supplies the energy for most active transport. 3. Electrogenic pump – a transport protein that generates voltage across a membrane; moves single proteins against an ion concentration gradient. |
|
|
What is the principal product of cellular respiration? Why is the production of this molecule important to cells?
|
1. The principle product of cellular respiration is ATP.
2. The production of ATP is important to cell because life on earth requires input of energy and photosynthesis provides almost all of the energy used by living organisms. |
|
|
Starting with a glucose molecule and 2 ATP, what are the carbon and energy outputs from glycolysis? What happens to the outputs from glycolysis in the absence of oxygen? In the presence of oxygen, what are the inputs into and outputs for the citric acid cycle? What are the inputs into and outputs from the electron transport chain? Overall, how many ATP are formed for each glucose molecule broken down in aerobic cellular respiration?
|
The carbon outputs from glycolysis are 2 molecules of pyruvate and the energy outputs are 2 net ATP and 2 NADH. Before the Citric Acid Cycle, the 2 molecules of pyruvate are converted to 2 molecules of acetyl CoA with the production of 2 molecules of CO2 and 2 molecules of NADH. Input into the Citric acid cycle is two molecules of acetyl CoA and the output is 4 molecules of CO2 2 ATP, 6 molecules of NADH and 2 molecules of FADH2. The inputs into the electron transport system are 10 molecules of NADH, 2 molecules of FADH2, and 6 oxygen molecules. The output from the electron transport system is 32 to 34 ATP. Approximately 36 to 38 ATP can be formed from a single molecule of glucose.
|
|
|
Where specifically is oxygen consumed in respiration? Where specifically is oxygen produced in photosynthesis?
|
1. In respiration, oxygen is specifically consumed in the mitochondrion.
2. In photosynthesis, oxygen is specifically consumed in the chloroplast. |
|
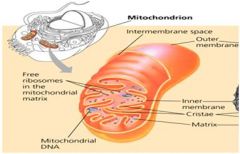
Draw a mitochondrion and distinguish among the inner and outer membranes, the intermembrane space and the matrix. Where specifically do glycolysis, the citric acid cycle and electron transport occur?
|
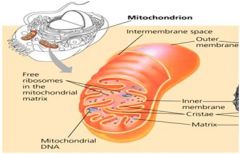
1. Glycolysis occurs in the cytoplasm.
2. The Citric Acid Cycle and Electron transport occur in the mitochondrion. |
|
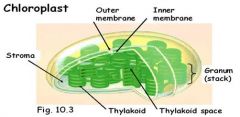
Draw a chloroplast. Identify the following structures or locations in the chloroplast: stroma, outer and inner membranes, thylakoid membranes, thylakoid space. Where are the components of the light reactions located? Where are the components of the Calvin cycle located? What pigment is most closely associated with photosynthesis and why is it green? What is fluorescence?
|
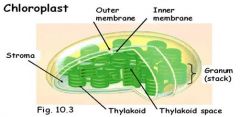
1. The components of the light reactions are located in the thylakoid membrane.
2. The components of the Calvin cycle are located in the stroma. 3. Photosynthesis is produced with wavelengths absorbed by chlorophyll; chlorophyll a & b absorb blue and red; not green. Leave appear to be green because green wavelengths are reflected or transmitted. 4. Fluorescence – when absorbed energy is reemitted as light energy of a longer wavelength. |
|
|
How many product cells are produced in mitosis and in meiosis? Relative to the starting mother cell, what is the genetic makeup of the cells produced in mitosis and in meiosis? What is synapsis? What is crossing over and when does it occur during meiosis?
|
1. Two daughter cells are formed with exactly the same chromosome composition is mitosis; meiosis II produces four daughter cells formed with half the number of chromosomes as the original mother cell.
2. Mitosis – independent alignment of chromosomes at metaphase; meiosis – homolygous chromosomes pair and separate in meiosis I. 3. Synapsis – the pairing of replicated homolygous chromosomes during prophase I of meiosis. 4. Crossing over – parts of chromatids exchanged producing genetic variation. |
|
|
Genetically, a cell can be described as 2n=46. What does "2n" stand for? What does the number 46 indicate about the cell?
|
2n=46 means the cells are diploid (2n) and in a diploid cell are 46 chromosomes, 2 sets of 23 homologous chromosomes. A sperm cell would possess 23 chromosomes since it should be haploid. (n=23).
|
|
|
Define the terms phenotype and genotype. Define and give examples of homozygous and heterozygous genotypes.
|
1. Phenotype – physical appearance; Genotype – genetic makeup.
2. Homozygous genotypes – consists of duplicates of the dominant or the recessive alleles (PP or pp). 3. Heterozygous genotypes – consists of two different alleles (Pp). |
|
|
Using the pea flower color trait studied by Mendel (purple and white flowers) give the genotype of the parental (P), true-breeding plants with purple and with white flowers. What kinds of gametes would each parent produce? What is the phenotype and genotype of the F1 offspring? If 2 F1 offspring are crossed to produce an F2 generation, what kind of gametes would each parent produce? What are the possible phenotypes and genotypes of the resulting F2 progeny and in what proportion would they be expected to be produced?
|

Answer in graph above.
|
|
|
Three different genes are considered at the same time in a cross (a trihybrid cross). Each gene follows Mendel's rules of inheritance. In this cross, E is dominant to e, F is dominant to f and G is dominant to g for the three traits. What is the probability of offspring possessing the dominant phenotype for each trait in the cross EeFfGg X EeFfGg?
|
1. Trihybrid Cross: EeFfGg x EeFfGg
1. Possible phenotypes – Ee x Ee, Ff x Ff, Gg x Gg 1.¾ x ¾ x ¾ = 27/64 2.Dominant phenotype (E_F_G_) – for EeFfGg 2n = 23 = 8. |
|
|
What is meant by incomplete dominance and codominance? A cross between snap dragon plants with red flowers and white flowers (P generation) produces pink flowers (F1 generation). What are the genotypes of the plants from these P and F1 generations? Two F1 individuals are then crossed. What gametes will be formed by the F1 individuals? What will be the phenotypes and genotypes of F2 individuals?
|
1. Incomplete dominance – heterozygous phenotype is intermediate between homozygous phenotypes.
2. Codominance – more than one type of allele can be dominant and expressed. 1. Genotypes: Red flowers – CRCR White flowers – CWCW Pink flowers – CRCW 2. P Generation – Red (CRCR) x White (CWCW) Gametes: CR x CW 3. F1 Generation – Pink (CRCW) Gametes: ½ CR x ½ CW 4. F2 Generation – CRCR, CRCW, CRCW, CWCW |
|
|
What is Huntington’s disease? How does Huntington’s disease differ from Cystic Fibrosis, Tay-Sachs and Sickle-Cell diseases in the way that it is inherited? Even though Huntington's disease is fatal it is still maintained in human populations. Why?
|
1. Huntington’s disease – a degenerative disease of the nervous system.
2. Huntington’s disease is caused by the lethal dominant allele that has no obvious phenotypic effect until the age of 35 to 45 years old. 3. The disease is produced by the dominant allele (no carriers, always expressed). 4. Huntington’s disease still remains today because people who are in a family history of Huntington’s disease have a 50% chance of inheriting the disease; and those that have it live to reproductive ages. |
|
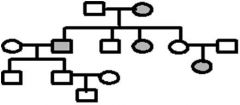
The pedigree below traces the inheritance of a rare biochemical disorder. Males are squares, females are circles and affected individuals are shaded. What is the best model for inheritance of this disorder? Fill in the genotypes of the individuals that are known from the pedigree information. What genotypes are possible for the other individuals
|
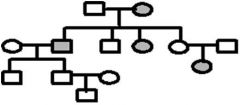
1. A pedigree is a diagram of a family tree showing the occurrence of heritable characters in parents and offspring over multiple generations. For a recessive trait, look to see if the trait skips generations in its inheritance. Skipping generations is typical if an individual must have the homozygous recessive condition for the trait to be expressed. Heterozygous individuals would be carriers and would not have the trait. If the trait is inherited as a Mendelian dominant it would be present in each generation.
2. White square – normal male 3. White circle – normal female 4. Grey square – infected male 5. Grey circle – inflected female |
|
|
Define the terms sex-linked genes and linked genes. Be able to distinguish between the two terms.
|
1. Sex-linked genes – trait is on the sex chromosome
2. Linked genes – genes located close enough together on a chromosome to be usually inherited together |
|
|
Hemophilia is a sex-linked trait on the X chromosome. Individuals with this genetic disorder fail to produce a clotting factor and may die when they receive minor cuts or even bruises. A normal male marries a normal female and they produce a son with hemophilia. What are the genotypes of these three individuals? What other genotypes and in what frequencies would you expect in all offspring from the original parents?
|
1. Mother genotype: XAXa
2. Father genotype: XAY 3. Son genotype: XaY 4. Females can be: XAXa, XaXa |
|
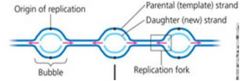
How are nucleotides bonded together to make a DNA strand? Explain what is meant by the description 5’ to 3’ (or 3’ to 5’) for a nucleic acid. What type of bonds form between pyrimidines and purines found in the nucleotides making up two DNA nucleic acids? What is the specific way that bases are paired when nucleic acid strands are bonded together? How is genetic information stored in a DNA molecule?
|
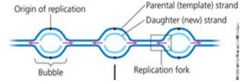
1. As individual nucleotides align with complementary nucleotides along a template strand of DNA, DNA polymerase adds them one by one, to the growing end of the new DNA strand.
2. 5’ >> 3’ means a new DNA strand elongates as the replication fork progresses. 3. 3’ >> 5’ means a lagging strand must grow by addition of short Okasaki segements. 4. Pyrimidines and purines form hydrogen bonds in base pairing DNA. 5. The specific way that bases are paired when nucleic acid strands are bonded together are by “ladders” twisted into a helix – one turn every 10 bases. 6. Genetic information is stored in DNA in the two nucleic acid strands within the sequence of nitrogenous bases. |
|
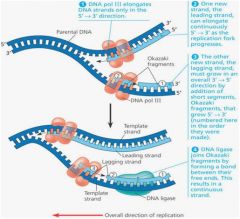
In DNA replication, what is a bubble, a point of origin, a replication fork? What is the general functions of the following enzymes: helicase, DNA pol III, primase, DNA pol I, and ligase? What does the abbreviation "pol" stand for? What are "leading" and "trailing" DNA strands? Why are separate mechanisms required for replication of these two types of strands? What do Okazaki fragments have to do with DNA replication?
|
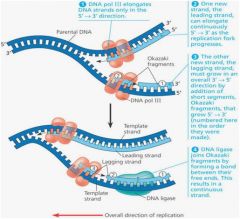
1. Semi-conservative refers to the fact that when a DNA molecule is replicated to form two new molecules, each new molecule contains one parental strand and one newly synthesized strand. A bubble is a portion of the DNA molecule that is untwisted and the hydrogen bonds between bases are broken separating a portion of the two strands. The point of origin is the center of the bubble and the replication fork is the point where strands are separating. The helicase enzyme separates the two DNA strands. DNA pol III, a DNA polymerase, reads a template strand and orders the correct complementary nucleotides to form the complementary DNA strand. Primase aligns RNA nucleotides to form a primer on a DNA template strand where DNA pol III will then bind and begin synthesizing the new daughter strand (adding nucleotides to the 3' end). DNA pol I replaces the RNA primer nucleotides with DNA nucleotides after the DNA pol III has finished its work. Ligase bonds pieces of the new DNA strand together. For example, after the DNA pol I has replaced the RNA primer nucleotides, the replacement nucleotides must be bonded to the growing daughter strand.
2. DNA replication is from the point of origin to the replication fork. One of the daughter strands being formed will be orientated 5' to 3', from the point of origin to the replication fork. This is a leading strand because the DNA pol III reads directly from the primer at the point of origin to the replication fork. Another daughter strand will read 3' to 5' from the point of origin to the replication fork. Since the DNA pol III can only add nucleotides to the 3' end of a strand it has to work in small stretches of DNA in a back word fashion. That is a primase establishes a primer several nucleotides toward the replication fork and the DNA pol II then synthesizes a segment of DNA from the primer back towards the point of origin. This is the lagging strand. The small stretches of DNA produced in this fashion on the lagging strand are termed the Okazaki fragments. Eventually the fragments are joined to make a continuous strand that overall grows from the point of origin to the replication fork. |
|
|
A three-nucleotide sequence of DNA or mRNA that specifies a particular amino acid or termination signal; the basic unit of the genetic code
|
codon
|
|
|
A type of RNA, synthesized from DNA, that attaches to ribosomes in the cytoplasm and specifies the primary structure of a protein.
|
messenger RNA (mRNA)
|
|
|
The premise that a gene is a segment of DNA that codes for one polypeptide.
|
one gene–one polypeptide hypothesis
|
|
|
An initial RNA transcript; also called pre-mRNA when transcribed from a protein-coding gene.
|
primary transcript
|
|
|
The way a cell’s mRNA-translating machinery groups the mRNA nucleotides into codons.
|
reading frame
|
|
|
A cell organelle constructed in the nucleolus and functioning as the site of protein synthesis in the cytoplasm; consists of rRNA and protein molecules, which make up two subunits.
|
ribosome
|
|
|
Modification of RNA before it leaves the nucleus, a process unique to eukaryotes.
|
RNA processing
|
|
|
The DNA strand that provides the template for ordering the sequence of nucleotides in an RNA transcript.
|
template strand
|
|
|
The synthesis of RNA on a DNA template.
|
transcription
|
|
|
The synthesis of a polypeptide using the genetic information encoded in an mRNA molecule. There is a change of languagefrom nucleotides to amino acids.
|
translation
|
|
|
A set of three-nucleotide-long words that specify the amino acids for polypeptide chains.
|
triplet code
|
|
|
A specific nucleotide sequence in DNA that binds RNA polymerase and indicates where to start transcribing RNA.
|
promoter
|
|
|
An enzyme that links together the growing chain of ribonucleotides during transcription.
|
RNA polymerase
|
|
|
A promoter DNA sequence crucial in forming the transcription initiation complex.
|
TATA box
|
|
|
In prokaryotes, a special sequence of nucleotides in DNA that marks the end of a gene. It signals RNA polymerase to release the newly made RNA molecule, which then departs from the gene.
|
terminator
|
|
|
A regulatory protein that binds to DNA and stimulates transcription of specific genes.
|
transcription factor
|
|
|
The completed assembly of transcription factors and RNA polymerase bound to the promoter.
|
transcription initiation complex
|
|
|
A region of a DNA molecule that is transcribed into an RNA molecule.
|
transcription unit
|
|
|
The 5' end of a pre-mRNA molecule modified by the addition of a cap of guanine nucleotide.
|
5' cap
|
|
|
A type of regulation at the RNA-processing level in which different mRNA molecules are produced from the same primary transcript, depending on which RNA segments are treated as exons and which as introns.
|
alternative RNA splicing
|
|
|
(1) A taxonomic category above the kingdom level. The three domains are Archaea, Bacteria, and Eukarya. (2) An independently folding part of a protein.
|
domain
|
|
|
A coding region of a eukaryotic gene. Exons, which are expressed, are separated from each other by introns.
|
exon
|
|
|
A noncoding, intervening sequence within a eukaryotic gene.
|
intron
|
|
|
The modified end of the 3’ end of an mRNA molecule consisting of the addition of some 50 to 250 adenine nucleotides.
|
poly-A tail
|
|
|
An enzymatic RNA molecule that catalyzes reactions during RNA splicing.
|
ribozyme
|
|
|
The removal of noncoding portions (introns) of the RNA molecule after initial synthesis.
|
RNA splicing
|
|
|
A complex assembly that interacts with the ends of an RNA intron in splicing RNA, releasing the intron and joining the two adjacent exons.
|
spliceosome
|
|
|
One of a ribosome’s three binding sites for tRNA during translation. The A site holds the tRNA carrying the next amino acid to be added to the polypeptide chain. (A stands for aminoacyl tRNA.)
|
A site
|
|
|
An enzyme that joins each amino acid to the correct tRNA.
|
aminoacyl-tRNA synthetase
|
|
|
A specialized base triplet at one end of a tRNA molecule that recognizes a particular complementary codon on an mRNA molecule.
|
anticodon
|
|
|
One of a ribosome’s three binding sites for tRNA during translation. The E site is the place where discharged tRNAs leave the ribosome. (E stands for exit.)
|
E site
|
|
|
One of a ribosome’s three binding sites for tRNA during translation. The P site holds the tRNA carrying the growing polypeptide chain. (P stands for peptidyl tRNA.)
|
P site
|
|
|
An aggregation of several ribosomes attached to one messenger RNA molecule.
|
polyribosome (polysome)
|
|
|
The most abundant type of RNA, which together with proteins forms the structure of ribosomes. Ribosomes coordinate the sequential coupling of tRNA molecules to mRNA codons.
|
ribosomal RNA (rRNA)
|
|
|
A stretch of amino acids on a polypeptide that targets the protein to a specific destination in a eukaryotic cell.
|
signal peptide
|
|
|
A protein-RNA complex that recognizes a signal peptide as it emerges from the ribosome.
|
signal-recognition particle (SRP)
|
|
|
An RNA molecule that functions as an interpreter between nucleic acid and protein language by picking up specific amino acids and recognizing the appropriate codons in the mRNA.
|
transfer RNA (tRNA)
|
|
|
A violation of the base-pairing rules in that the third nucleotide (5’ end) of a tRNA anticodon can form hydrogen bonds with more than one kind of base in the third position (3’ end) of a codon.
|
wobble
|
|
|
A type of point mutation; the replacement of one nucleotide and its partner in the complementary DNA strand by another pair of nucleotides.
|
base-pair substitution
|
|
|
(1) A deficiency in a chromosome resulting from the loss of a fragment through breakage. (2) A mutational loss of one or more nucleotide pairs from a gene.
|
deletion
|
|
|
A mutation occurring when the number of nucleotides inserted or deleted is not a multiple of three, resulting in the improper grouping of the following nucleotides into codons.
|
frameshift mutation
|
|
|
A mutation involving the addition of one or more nucleotide pairs to a gene.
|
insertion
|
|
|
The most common type of mutation, a base-pair substitution in which the new codon makes sense in that it still codes for an amino acid.
|
missense mutation
|
|
|
A chemical or physical agent that interacts with DNA and causes a mutation
|
mutagen
|
|
|
A rare change in the DNA of a gene, ultimately creating genetic diversity.
|
mutation
|
|
|
A mutation that changes an amino acid codon to one of the three stop codons, resulting in a shorter and usually nonfunctional protein.
|
nonsense mutation
|
|
|
A change in a gene at a single nucleotide pair.
|
point mutation
|
|
|
The name of the late stages of HIV infection, defined by a specified reduction of T cells and the appearance of characteristic secondary infections.
|
AIDS (acquired immunodeficiency syndrome)
|
|
|
A virus that infects bacteria; also called a phage.
|
bacteriophage
|
|
|
The protein shell that encloses a viral genome. It may be rod-shaped, polyhedral, or more complex in shape.
|
capsid
|
|
|
The infectious agent that causes AIDS. HIV is a retrovirus.
|
HIV (human immunodeficiency virus)
|
|
|
The limited range of host cells that each type of virus can infect and parasitize.
|
host range
|
|
|
A phage replication cycle in which the viral genome becomes incorporated into the bacterial host chromosome as a prophage and does not kill the host.
|
lysogenic cycle
|
|
|
A type of viral (phage) replication cycle resulting in the release of new phages by lysis (and death) of the host cell.
|
lytic cycle
|
|
|
A virus that infects bacteria; also called a bacteriophage.
|
phage
|
|
|
A phage genome that has been inserted into a specific site on the bacterial chromosome.
|
prophage
|
|
|
Viral DNA that inserts into a host genome.
|
provirus
|
|
|
An RNA virus that reproduces by transcribing its RNA into DNA and then inserting the DNA into a cellular chromosome; an important class of cancer-causing viruses.
|
retrovirus
|
|
|
An enzyme encoded by some certain viruses (retroviruses) that uses RNA as a template for DNA synthesis.
|
reverse transcriptase
|
|
|
A membrane that cloaks the capsid that in turn encloses a viral genome.
|
viral envelope
|
|
|
In prokaryotes, the direct transfer of DNA between two cells that are temporarily joined. In ciliates, a sexual process in which two cells exchange haploid micronuclei.
|
conjugation
|
|
|
A small ring of DNA that carries accessory genes separate from those of a bacterial chromosome; also found in some eukaryotes, such as yeast.
|
plasmid
|
|
|
(1) A DNA transfer process in which phages carry bacterial genes from one host cell to another. (2) In cellular communication, the conversion of a signal from outside the cell to a form that can bring about a specific cellular response.
|
transduction
|
|
|
(1) The conversion of a normal animal cell to a cancerous cell. (2) A change in genotype and phenotype due to the assimilation of external DNA by a cell.
|
transformation
|
|
|
A segment of DNA that can move within the genome of a cell by means of a DNA or RNA intermediate; also called a transposable element.
|
transposable genetic element
|
|
|
A transposable genetic element that moves within a genome by means of a DNA intermediate.
|
transposon
|
|
|
A protein that binds to DNA and stimulates transcription of a specific gene.
|
activator
|
|
|
A small molecule that cooperates with a repressor protein to switch on operon off.
|
corepressor
|
|
|
Cyclic adenosine monophosphate, a ring-shaped molecule made from ATP that is a common intracellular signaling molecule (second messenger) in eukaryotic cells (for example, in vertebrate endocrine cells). It is also a regulator of some bacterial operons.
|
cyclic AMP (cAMP)
|
|
|
A specific small molecule that inactivates the repressor in an operon.
|
inducer
|
|
|
In prokaryotic DNA, a sequence of nucleotides near the start of an operon to which an active repressor can attach. The binding of the repressor prevents RNA polymerase from attaching to the promoter and transcribing the genes of the operon.
|
operator
|
|
|
A unit of genetic function common in bacteria and phages, consisting of coordinately regulated clusters of genes with related functions.
|
operon
|
|
|
A bacterial plasmid carrying genes that confer resistance to certain antibiotics.
|
R plasmid
|
|
|
A gene that codes for a protein, such as a repressor, that controls the transcription of another gene or group of genes.
|
regulatory gene
|
|
|
A protein that suppresses the transcription of a gene.
|
repressor
|
|
|
The structural and functional divergence of cells as they become specialized during a multicellular organism’s development; dependent on the control of gene expression.
|
cell differentiation
|
|
|
A segment of noncoding DNA that helps regulate transcription of a gene by binding proteins called transcription factors.
|
control element
|
|
|
The expression of different sets of genes by cells with the same genome.
|
differential gene expression
|
|
|
A DNA segment containing multiple control elements that may be located far away from the gene it regulates.
|
enhancer
|
|
|
The attachment of acetyl groups to certain amino acids of histone proteins.
|
histone acetylation
|
|
|
A protein that suppresses the transcription of a gene.
|
repressor
|
|
|
A technique to silence the expression of selected genes in nonmammalian organisms. The method uses synthetic double-stranded RNA molecules matching the sequence of a particular gene to trigger the breakdown of the gene’s messenger RNA.
|
RNA interference (RNAi)
|
|
|
A regulatory protein that binds to DNA and stimulates transcription of specific genes.
|
transcription factor
|
|
|
A collection of genes with similar or identical sequences, presumably of common origin.
|
multigene family
|
|
|
A DNA segment very similar to a real gene but which does not yield a functional product; a gene that has become inactivated in a particular species because of mutation.
|
pseudogene
|
|
|
Nucleotide sequences, usually noncoding, that are present in many copies in a eukaryotic genome. The repeated units may be short and arranged tandemly (in series) or long and dispersed in the genome.
|
repetitive DNA
|
|
|
A transposable element that moves within a genome by means of an RNA intermediate, a transcript of the retrotransposon DNA.
|
retrotransposon
|
|
|
A transposable genetic element that moves within a genome by means of a DNA intermediate.
|
transposon
|
|
|
The manipulation of living organisms or their components to produce useful products.
|
biotechnology
|
|
|
A limited gene library using complementary DNA. The library includes only the genes that were transcribed in the cells examined.
|
cDNA library
|
|
|
(1) A lineage of genetically identical individuals or cells. (2) In popular usage, a single individual organism that is genetically identical to another individual. (3) As a verb, to make one or more genetic replicas of an individual or cell. See also gene cloning.
|
clone
|
|
|
An agent used to transfer DNA in genetic engineering. A plasmid that moves recombinant DNA from a test tube back into a cell is an example of a cloning vector, as is a virus that transfers recombinant DNA by infection.
|
cloning vector
|
|
|
A DNA molecule made in vitro using mRNA as a template and the enzyme reverse transcriptase. A cDNA molecule therefore corresponds to a gene, but lacks the introns present in the DNA of the genome.
|
complementary DNA (cDNA)
|
|
|
A cloning vector that contains the requisite prokaryotic promoter just upstream of a restriction site where a eukaryotic gene can be inserted.
|
expression vector
|
|
|
The production of multiple copies of a gene.
|
gene cloning
|
|
|
The direct manipulation of genes for practical purposes.
|
genetic engineering
|
|
|
A set of thousands of DNA segments from a genome, each carried by a plasmid, phage, or other cloning vector.
|
genomic library
|
|
|
A DNA molecule made in vitro with segments from different sources.
|
recombinant DNA
|
|
|
A degradative enzyme that recognizes and cuts up DNA (including that of certain phages) that is foreign to a bacterium.
|
restriction enzyme
|
|
|
DNA segment resulting from cutting of DNA by a restriction enzyme.
|
restriction fragment
|
|
|
A specific sequence on a DNA strand that is recognized as a cut siteby a restriction enzyme.
|
restriction site
|
|
|
A single-stranded end of a double-stranded DNA restriction fragment.
|
sticky end
|
|
|
The study of whole sets of genes and their interactions.
|
genomics
|
|
|
A technique to discover the function of a gene by introducing specific changes into the sequence of a cloned gene, reinserting the mutated gene into a cell, and studying the phenotype of the mutant.
|
in vitro mutagenesis
|
|
|
The systematic study of the full protein sets (proteomes) encoded by genomes.
|
proteomics
|
|
|
A technique to silence the expression of selected genes in nonmammalian organisms. The method uses synthetic double-stranded RNA molecules matching the sequence of a particular gene to trigger the breakdown of the gene’s messenger RNA.
|
RNA interference (RNAi)
|
|
|
The alteration of the genes of a person afflicted with a genetic disease.
|
gene therapy
|
|
|
An organism that has acquired one or more genes by artificial means; also known as a transgenic organism.
|
genetically modified (GM) organism
|
|
|
In multicellular organisms, one of many types of circulating chemical signals that are formed in specialized cells, travel in body fluids, and act on specific target cells to change their functioning
|
hormone
|
|
|
A chemical messenger that influences cells in the vicinity.
|
local regulator
|
|
|
In cellular communication, the target cell’s detection (by binding to a receptor protein) of a signal molecule from outside the cell.
|
reception
|
|
|
In cellular communication, the change in a specific cellular activity brought about by a transduced signal from outside the cell.
|
response
|
|
|
A mechanism linking a mechanical or chemical stimulus to a specific cellular response.
|
signal transduction pathway
|
|
|
A DNA transfer process in which phages carry bacterial genes from one host cell to another. (2) In cellular communication, the conversion of a signal from outside the cell to a form that can bring about a specific cellular response.
|
transduction (1)
|
|
|
A GTP-binding protein that relays signals from a plasma membrane signal receptor, known as a G-protein-linked receptor, to other signal transduction proteins inside the cell. When such a receptor is activated, it in turn activates the G protein, causing it to bind a molecule of GTP in place of GDP. Hydrolysis of the bound GTP to GDP inactivates the G protein
|
G protein
|
|
|
A signal receptor protein in the plasma membrane that responds to the binding signal molecule by activating a G protein.
|
G-protein-linked receptor
|
|
|
A molecule that binds specifically to a receptor site of another molecule.
|
ligand
|
|
|
A protein pore in the plasma membrane that opens or closes in response to a chemical signal, allowing or blocking the flow of specific ions.
|
ligand-gated ion channel
|
|
|
A receptor protein in the plasma membrane that responds to the binding of a signal molecule by catalyzing the transfer of phosphate groups from ATP to tyrosines on the cytoplasmic side of the receptor. The phosphorylated tyrosines activate other signal transduction proteins within the cell.
|
receptor tyrosine kinase
|
|
|
An enzyme that catalyzes the transfer of phosphate groups from ATP to the amino acid tyrosine on a substrate protein.
|
tyrosine kinase
|
|
|
An enzyme that converts ATP to cyclic AMP in response to a chemical signal.
|
adenylyl cyclase
|
|
|
Cyclic adenosine monophosphate, a ring-shaped molecule made from ATP that is a common intracellular signaling molecule (second messenger) in eukaryotic cells (for example, in vertebrate endocrine cells). It is also a regulator of some bacterial operons.
|
cyclic AMP (cAMP)
|
|
|
A second messenger that functions as an intermediate between certain nonsteroid hormones and a third messenger, a rise in cytoplasmic Ca2+ concentration.
|
inositol trisphosphate (IP3)
|
|
|
An enzyme that transfers phosphate groups from ATP to a protein.
|
protein kinase
|
|
|
An enzyme that removes phosphate groups from proteins, often functioning to reverse the effect of a protein kinase.
|
protein phosphatase
|
|
|
A small, nonprotein, water-soluble molecule or ion, such as calcium ion or cyclic AMP, that relays a signal to a cell’s interior in response to a signal received by a signal receptor protein.
|
second messenger
|
|
|
Nonliving.
|
abiotic
|
|
|
A plant hormone that slows down growth, often antagonizing actions of growth hormones. Two of its many effects are to promote seed dormancy and facilitate drought tolerance.
|
abscisic acid (ABA)
|
|
|
The changes that occur within a cell as it undergoes programmed cell death, which is brought about by signals that trigger the activation of a cascade of suicide proteins in the cell destined to die.
|
apoptosis
|
|
|
A term that primarily refers to indoleacetic acid (IAA), a natural plant hormone that has a variety of effects, including cell elongation, root formation, secondary growth, and fruit growth.
|
auxin
|
|
|
Referring to all the organisms that are part of the environment.
|
biotic
|
|
|
Steroid hormones in plants that have a variety of effects, including cell elongation, retarding leaf abscission, and promoting xylem differentiation.
|
brassinosteroids
|
|
|
A class of related plant hormones that retard aging and act in concert with auxin to stimulate cell division, influence the pathway of differentiation, and control apical dominance.
|
cytokinins
|
|
|
The only gaseous plant hormone. Among its many effects are response to mechanical stress, programmed cell death, leaf abscission, and fruit ripening.
|
ethylene
|
|
|
Plant enzymes that break the cross-links (hydrogen bonds) between cellulose microfibrils and other cell wall constituents, loosening the wall’s fabric.
|
expansins
|
|
|
A class of related plant hormones that stimulate growth in the stem and leaves, trigger the germination of seeds and breaking of bud dormancy, and stimulate fruit development with auxin.
|
gibberellins
|
|
|
In multicellular organisms, one of many types of circulating chemical signals that are formed in specialized cells, travel in body fluids, and act on specific target cells to change their functioning.
|
hormone
|
|
|
Growth of a plant shoot toward or away from light.
|
phototropism
|
|
|
A plant growth maneuver in response to mechanical stress, involving slowing of stem elongation, a thickening of the stem, and a curvature that causes the stem to start growing horizontally.
|
triple response
|
|
|
A growth response that results in the curvature of whole plant organs toward or away from stimuli owing to differential rates of cell elongation.
|
tropism
|
|
|
One of two endocrine glands located adjacent to the kidneys in mammals. Endocrine cells in the outer portion (cortex) respond to ACTH by secreting steroid hormones that help maintain homeostasis during long-term stress. Neurosecretory cells in the central portion (medulla) secrete epinephrine and norepinephrine in response to nervous inputs triggered by short-term stress.
|
adrenal gland
|
|
|
A hormone produced in the hypothalamus and released from the posterior pituitary. It promotes water rentention by the kidneys as part of an elaborate feedback scheme that helps regulate the osmolarity of the blood.
|
antidiuretic hormone (ADH)
|
|
|
A hormone secreted by the thyroid gland that lowers blood calcium levels by promoting calcium deposition in bone and calcium excretion from the kidneys.
|
calcitonin
|
|
|
An endocrine disorder marked by inability to maintain glucose homeostasis. The type I form results from autoimmune destruction of insulin-secreting cells; treatment usually requires insulin injections several times a day. The type II form most commonly results from reduced responsiveness of target cells to insulin; obesity and lack of exercise are risk factors.
|
diabetes mellitus
|
|
|
Any of several hormones produced in the brain and anterior pituitary that inhibits pain perception.
|
endorphin
|
|
|
A hormone secreted by pancreatic alpha cells that raises blood glucose levels. It promotes glycogen breakdown and release of glucose by the liver.
|
glucagon
|
|
|
The ventral part of the vertebrate forebrain; functions in maintaining homeostasis, especially in coordinating the endocrine and nervous systems; secretes hormones of the posterior pituitary and releasing factors that regulate the anterior pituitary.
|
hypothalamus
|
|
|
A hormone secreted by pancreatic beta cells that lowers blood glucose levels. It promotes the uptake of glucose by most body cells and the synthesis and storage of glycogen in the liver and also stimulates protein and fat synthesis.
|
insulin
|
|
|
Clusters of endocrine cells within the pancreas that produce and secrete the hormones glucagon (alpha cells) and insulin (beta cells).
|
islets of Langerhans
|
|
|
A gland with dual functions: The nonendocrine portion secretes digestive enzymes and an alkaline solution into the small intestine via a duct; the endocrine portion secretes the hormones insulin and glucagon into the blood.
|
pancreas
|
|
|
Any of four small endocrine glands, embedded in the surface of the thyroid gland, that secrete parathyroid hormone.
|
parathyroid gland
|
|
|
A hormone secreted by the parathyroid glands that raises blood calcium level by promoting calcium release from bone and calcium retention by the kidneys.
|
parathyroid hormone (PTH)
|
|
|
An endocrine gland at the base of the hypothalamus; consists of a posterior lobe (neurohypophysis), which stores and releases two hormones produced by the hypothalamus, and an anterior lobe (adenohypophysis), which produces and secretes many hormones that regulate diverse body functions.
|
pituitary gland
|
|
|
Also called the neurohypophysis; an extension of the hypothalamus composed of nervous tissue that secretes oxytocin and antidiscretic hormone made in the hypothalamus; a temporary storage site for these hormones.
|
posterior pituitary
|
|
|
An endocrine gland, located on the ventral surface of the trachea, that secretes two iodine-containing hormones, triiodothyronine (T3) and thyroxine (T4), and cacitonin.
|
thyroid gland
|
|
|
One of two iodine-containing hormones that are secreted by the thyroid gland and help regulate metabolism, development, and maturation in vertebrates.
|
thyroxine (T4)
|
|
|
One of two iodine-containing hormones that are secreted by the thyroid gland and help regulate metabolism, development, and maturation in vertebrates.
|
triiodothyrodine (T3)
|
|
|
A hormone that has another endocrine gland as a target.
|
tropic hormone
|
|
|
One of the fat-soluble vitamins. The active form functions as a hormone, acting in concert with parathyroid hormone in bone and promoting the uptake of calcium from food within the intestines.
|
vitamin D
|
|
|
A typically long extension, or process, from a neuron that carries nerve impulses away from the cell body toward target cells.
|
axon
|
|
|
The conical region of a neuron’s axon where it joins the cell body; typically the region where nerve signals are generated.
|
axon hillock
|
|
|
The part of a cell, such as a neuron, that houses the molecules.
|
cell body
|
|
|
In vertebrate animals, the brain and spinal cord.
|
central nervous system (CNS)
|
|
|
One of usually numerous, short, highly branched processes of a neuron that convey nerve impulses toward the cell body.
|
dendrite
|
|
|
A muscle cell or gland cell that performs the body’s responses to stimuli; responds to signals from the brain or other processing center of the nervous system.
|
effector cell
|
|
|
An association neuron; a nerve cell within the central nervous system that forms synapses with sensory and motor neurons and integrates sensory input and motor output.
|
interneuron
|
|
|
A nerve cell that transmits signals from the brain or spinal cord to muscles or glands.
|
motor neuron
|
|
|
In a neuron, an insulating coat of cell membrane from Schwann cells that is interrupted by nodes of Ranvier, where saltatory conduction occurs.
|
myelin sheath
|
|
|
A chemical messenger released from the synaptic terminal of a neuron at a chemical synapse that diffuses across the synaptic cleft and binds to and stimulates the postsynaptic cell.
|
neurotransmitter
|
|
|
A type of glial cell that forms insulating myelin sheaths around the axons of neurons in the central nervous system.
|
oligodendrocyte
|
|
|
The sensory and motor neurons that connect to the central nervous system.
|
peripheral nervous system (PNS)
|
|
|
The target cell at a synapse.
|
postsynaptic cell
|
|
|
The transmitting cell at a synapse.
|
presynaptic cell
|
|
|
A type of glial cells that forms insulating myelin sheaths around the axons of neurons in the peripheral nervous system.
|
Schwann cell
|
|
|
A nerve cell that receives information from the internal and external environments and transmits the signals to the central nervous system.
|
sensory neuron
|
|
|
The locus where one neuron communicates with another neuron in a neural pathway; a narrow gap between a synaptic terminal of an axon and a signal-receiving portion (dendrite or cell body) of another neuron or effector cell. Neurotransmitter molecules released by synaptic terminals diffuse across the synapse, relaying messages to the dendrite or effector.
|
synapse
|
|
|
A bulb at the end of an axon in which neurotransmitter molecules are stored and released.
|
synaptic terminal
|
|
|
The magnitude of a cell’s membrane voltage at equilibrium; calculated using the Nernst equation.
|
equilibrium potential (Eion)
|
|
|
A gated channel for a specific ion. By opening and closing such channels, a cell alters its membrane potential.
|
gated ion channel
|
|
|
A protein pore in the plasma membrane that opens or closes in response to a chemical signal, allowing or blocking the flow of specific ions.
|
ligand-gated ion channel
|
|
|
The charge difference between a cell’s cytoplasm and the extracellular fluid, due to the differential distribution of ions. Membrane potential affects the activity of excitable cells and the transmembrane movement of all charged substances.
|
membrane potential
|
|
|
The membrane potential characteristic of a nonconducting, excitable cell, with the inside of the cell more negative than the outside.
|
resting potential
|
|
|
A specialized ion channel that opens or closes in response to changes in membrane potential.
|
voltage-gated ion channel
|
|
|
A rapid change in the membrane potential of an excitable cell, caused by stimulus-triggered, selective opening and closing of voltage-sensitive gates in sodium and potassium ion channels.
|
action potential
|
|
|
An electrical state in an excitable cell whereby the inside of the cell is made less negative relative to the outside than at the resting membrane potential. A neuron membrane is depolarized if a stimulus decreases its voltage from the resting potential of -70 mV in the direction of zero voltage.
|
depolarization
|
|
|
A local voltage change in a neuron membrane induced by stimulation of a neuron, with strength proportional to the strength of the stimulus and lasting about a millisecond.
|
graded potential
|
|
|
An electrical state whereby the inside of the cell is made more negative relative to the outside than at the resting membrane potential. A neuron membrane is hyperpolarized if a stimulus increases its voltage from the resting potential of -70 mV, reducing the chance that the neuron will transmit a nerve impulse.
|
hyperpolarization
|
|
|
The potential an excitable cell membrane must reach for an action potential to be initiated.
|
threshold
|
|
|
One of the most common neurotransmitters; functions by binding to receptors and altering the permeability of the postsynaptic membrane to specific ions, either depolarizing or hyperpolarizing the membrane.
|
acetylcholine
|
|
|
Any of several hormones produced in the brain and anterior pituitary that inhibits pain perception
|
endorphin
|
|
|
A catecholamine hormone secreted from the adrenal medulla that mediates fight-or-flightresponses to short-term stress; also functions as a neurotransmitter.
|
epinephrine
|
|
|
An electrical change (depolarization) in the membrane of a postsynaptic neuron caused by the binding of an excitatory neurotransmitter from a presynaptic cell to a postsynaptic receptor; makes it more likely for a postsynaptic neuron to generate an action potential.
|
excitatory postsynaptic potential (EPSP)
|
|
|
An electrical charge (hyperpolarization) in the membrane of a postsynaptic neuron caused by the binding of an inhibitory neurotransmitter from a presynaptic cell to a postsynaptic receptor; makes it more difficult for a postsynaptic neuron to generate an action potential.
|
inhibitory postsynaptic potential (IPSP)
|
|
|
A relatively short chain of amino acids that serves as a neurotransmitter.
|
neuropeptide
|
|
|
A phenomenon of neural integration in which the membrane potential of the postsynaptic cell is determined by the combined effect of EPSPs or IPSPs produced nearly simultaneously by different synapses.
|
spatial summation
|
|
|
A narrow gap separating the synaptic knob of a transmitting neuron from a receiving neuron or an effector cell.
|
synaptic cleft
|
|
|
Membranous sac containing neurotransmitter molecules at the tip of the presynaptic axon.
|
synaptic vesicle
|
|
|
A phenomenon of neural integration in which the membrane potential of the postsynaptic cell in a chemical synapse is determined by the combined effect of EPSPs or IPSPs produced in rapid succession.
|
temporal summation
|
|
|
What organism did Beadle and Tatum use to study the relationships between genes and gene products? What general class of mutants did they study and how did they obtain these mutants? What did their results tell them about arginine synthesis in their experimental organism? What did Beadle and Tatum conclude about the relationship between arginine synthesis and gene products?
|
Beadle and Tatum studied a bread mold, Neurospora sp. They studied mutants that unlike wild type Neurospora were unable to synthesize the amino acid arginine and therefore required this amino acid for growth. The mutants were obtained by exposing Neurospora to X-rays. By examining the growth of the arginine mutants on different compounds, they were first able to determine the pathway by which arginine was synthesized and the surmised that their mutants were deficient in single steps of the pathway. Beadle and Tatum concluded that their mutants were deficient in individual enzymes and proposed that single genes coded for single enzymes.
|
|
|
How has our present concept of a gene and its specific product been refined since Beadle and Tatum?
|
Based on the facts that not all proteins are enzymes and that some enzymes are composed of more than one type of polypeptide it can more precisely be that one gene codes for one polypeptide.
|
|
|
Why is the genetic code referred to as a triplet code? What is a codon? Why do we say the code is specific but redundant? What does the codon AUG code for? Why do we describe the genetic code as “universal”? How does the tobacco plant transformed with firefly luciferase illustrate you answer?
|
The genetic code is called a triplet code because a single amino acid is coded for by the sequence of three bases. A codon is term applied to the mRNA sequence that produces a specific amino acid. The code is considered specific because a codon codes for only one specific amino acid. The code is termed redundant because a single amino acid may be coded for by up to four different three-base sequences. The codon AUG codes for the amino acid methionine and is recognized as a "start" codon, the codon where translation should begin on an mRNA. The genetic code is universal in that it is virtually the same code in all living organisms. In the tobacco plant the genetic code for luciferase from fire flies is recognized and correctly translated in to the luciferase protein by the tobacco plant.
|
|
|
Be able to determine the amino acid a codon produces using Fig. 17.5. For example, what does GCC code for? What possible codons produce the amino acid threonine (Thr)? What amino acid would be produced by the DNA sequence 3’-GCG-5’? (Hint: convert this sequence to a codon and then determine the amino acid.)
|
The mRNA codon GCC codes for the amino acid alanine. Threonine is produced by ACU, ACC, ACA and ACG. The DNA sequence GCG would produce the mRNA codon CGC which codes for the amino acid arginine (Arg).
|
|
|
The base sequence on a stretch of DNA is 3’-ACCAAACCGACT-5’. What would be the corresponding sequence on a mRNA made from this DNA? What would the sequence of amino acids that would be made from the mRNA formed by the original DNA (you will need Fig. 17.5)?
|
The mRNA would be UGGUUUGGCUGA. The sequence of amino acids would be trp-phe-gly (UGA is a stop sequence and it does not code for an amino acid).
|
|
|
What does transcription produce and where does it occur in a eukaryotic cell?
|
Transcription produces an RNA molecule. In an eukaryotic cell, transcription produces a pre-mRNA which must be processed in two steps to produce true mRNA.
|
|
|
What are the three general steps or phases of transcription?
|
Transcription can be divided into an initiation phase. an elongation phase and a termination phase.
|
|
|
What is a promoter and what does it have to do with the initiation of transcription? What is the direction that a DNA template is read in transcription? What is the direction that the corresponding RNA is formed?
|
A promoter is a sequence of DNA that is upstream from a gene. It is the site where an RNA polymerase molecule binds and it initiates the transcription of a protein. The template strand of DNA is read 3' to 5' and the mRNA is formed in the opposite orientation, 5' to 3'.
|
|
|
What is RNA processing? Specifically, what are the two steps in which a pre-mRNA is converted to an mRNA in a eukaryotic cell? Describe what is involved in each step and the apparent function of each step. Structurally, how does quanosine triphosphate differ in structure from adenosine triphosphate.
|
RNA processing is the modification of an mRNA after transcription in a eukaryotic cell. The first step is the modification of the ends of the RNA molecule. A molecule of guanosine triphosphate added to the 5' end of the RNA molecule and 50 to 250 adenine nucleotides are added to the 3' end of the molecule. In the second step, enzymes cut out portions of the mRNA called introns which do not code for proteins and the remaining exons (which do code for proteins) are spliced together. Guanosine triphosphate is a nucleotide that has the same structure as ATP except the adenine base is replaced by guanine.
|
|
|
Where does translation occur in a eukaryotic cell and what is the product of translation?
|
Translation occurs in the cytoplasm of a eukaryotic cell and the product is a polypeptide.
|
|
|
Describe the general function of tRNA, aminoacyl-tRNA synthetase, ribosomes. What is the general structure of a tRNA and a ribosome? Where are these molecules synthesized in a eukaryotic cell.
|
tRNA (transfer RNA) is an RNA molecule that brings specific amino acids to ribosomes where polypeptides are being synthesized. The aminoacyl-tRNA synthetase catalyzes the addition of specific amino acids to tRNAs for synthesis of polypeptides during translation. A ribosome is a complex that is structured from protein and RNA. Ribosomes are the site of translation. A tRNA molecule is a single strand of RNA that is about 80 bases long. Certain bases in the molecule complement one another so that it folds on itself. At the 3' end of the molecule is a site of attachment for specific amino acids and at the other end of the folded molecule is specific 3 base sequence, the anticodon, which recognizes specific codons on mRNA. Ribosomes are formed from a large and a small subunit which assemble during translation on a mRNA molecule. Both mRNA and ribosomes are synthesized in the nucleus of eukaryotic cells. Ribosomes are synthesized from RNA and proteins in nucleoli. The ribosome proteins are produced in the cytoplasm and imported into the nucleoli.
|
|
|
How is translation initiated in a eukaryotic cell? After initiation, what happens to cause elongation of a polypeptide? What causes termination of translation and what happens to mRNA and ribosomes after termination?
|
Translation is initiated when an mRNA combines with a small subunit of a ribosome and an initiator tRNA then a large ribosomal subunit. After initiation, the ribosome moves down the mRNA and a tRNA (attached to a specific amino acid) that corresponds to the next codon binds to the ribosome at its A site (aminoacyl-tRNA binding site). The amino acid at the A site binds to the growing polypeptide that is attached to the tRNA occupying the P (peptidyl-tRNA binding) site. The tRNA that now is attached to the growing polypeptide moves to the P site, the tRNA that occupied the P site moves to the E (exit) site on the ribosome and a new tRNA matching the next codon on the RNA binds to the A site. When a ribosome reaches a stop codon on the mRNA, the A site is occupied by a protein called release factor instead of another tRNA. The release factor acts to break the bond between the growing polypeptide and the tRNA occupying the P site on the ribosome. Breaking this bond releases the polypeptide and stimulates disassembly of the ribosome.
|
|
|
What is a polyribosome? How do polyribosomes effect protein synthesis in a cell?
|
A polyribosome is made up of several ribosomes on translating the same mRNA. Polyribosomes are a mechanism to produce multiple copies of a protein in a short period of time.
|
|
|
What are three ways that a polypeptide may change to become a functional protein? Briefly describe what happens in each type of change.
|
A polypeptide may be 1) folded to form the functional 3-dimensional structure; 2) Modified by attachment of sugars or lipids, removal or addition of amino acids after translation, or cleaved into parts or assembled with other polypeptides; 3) targeted and transported to sites of function that are separate from the cytoplasm.
|
|
|
Describe how transcription and translation can be coupled in prokaryotic cells. What is the advantage of coupling transcription and translation in a prokaryotic cell? Why are transcription and translation not as directly coupled in eukaryotic cells?
|
In prokaryotic cells, ribosomes can begin translating a mRNA before it is completed so that as the mRNA is being formed, the 5' end is already being translated before transcription is complete. Further, more than one ribosome can translate the same mRNA so that while the mRNA is being formed, several polypeptides can be formed in translation. Coupling provides an mechanism by which many copies of a prokaryotic protein can be rapidly formed. Transcription occurs in the nucleus of eukaryotic cells while translation occurs in the cytoplasm. This separation precludes coupling of the two processes.
|
|
|
What is a "point mutation" and do they occur in DNA or RNA? How are missense and nonsense mutations different? Sickle-cell disease is an example of what can happen with a base pair substitution (a point mutation). How does this substitution change the normal gene product? What does the term "frameshift" mean with regard to RNA transcription? How can a deletion or insertion (also types of point mutations) cause a frameshift mutation?
|
Point mutations are alterations in DNA where there is substitution, loss or addition of a single nucleotide. In sickle-cell disease, the substitution of a single base results in the replacement of the amino acid glutamic acid with valine. These two amino acids have quite different properties and as a result, tertiary properties of the polypeptide and the quaternary structure of hemoglobin is altered leading to miss-shaped red blood cells and a host of physiological consequences. A point mutation that adds or subtracts a nucleotide will shift the order of nucleotides that make up the codons in an mRNA. For example, three consecutive codons read UUUGCACGA, which translates to phenylalanine, alanine, and arginine. If a nucleotide deletion removes the 2nd base, U, then the ribosomes will read UUGCACGA. UUG codes for leucine, histidine for the first two amino acids.
|
|
|
What is the approximate size of a virus? How does this compare to the sizes of bacterial and eukaryotic cells? We call viruses obligate intracellular parasites. What does this mean?
|
Viruses usually range in size from 0.05 to 0.2 µm. By comparison bacteria range from 1 to 10 µm and eukaryotic cells range from 10 - 100 µm. The size of viruses is approximately 1/20th that of bacterial or prokaryotic cells. Viruses are obligate parasites because they can only survive and be reproduced in their host cells.
|
|
|
What are two structural components shared by all viruses? What other structures can viruses possess?
|
All viruses possess a genome in the form of either an RNA molecule or a DNA molecule surrounded by a protein structure termed a capsid. Some viruses possess a viral envelope or membrane and specific glycoproteins anchored in this membrane.
|
|
|
What are the different types of genomes possessed by viruses? What is probably the functional reason for these different types of genomes?
|
Viral genomes can be double-stranded DNA, single-stranded DNA, double stranded RNA or single stranded RNA. In the host, the single-stranded RNA can act as a mRNA, a template for a mRNA or a template for DNA, depending on the specific virus. These forms probably reflect ways that viral genomes evolved to avoid the defenses of their host cells.
|
|
|
What kind of cells does HIV target and how does infection occur? What is reverse transcriptase and what is its role in reproduction of HIV? How do infected HIV particles leave infected host cells?
|
HIV (human immunodeficiency virus target T-cells, specific types of macrophages or white blood cells. HIV infection occurs recognition and binding of glycoproteins in the viral envelope with receptors on the membrane of the T-cells. The virus and T-cell membrane come in contact and fuse and the viral particle is engulfed into the host cell. Reverse transcriptase in an enzyme that catalyzes the formation of a DNA strand using the HIV RNA as a template. The DNA from the viral template is transported into the host cell nucleus and inserted into the host cell DNA. The HIV DNA is used to make RNA copies that include viral genomes and other RNA molecules that code for proteins required by the virus including the capsid proteins, reverse transcriptase and glycoproteins for the viral envelope. HIV genome molecules, capsid molecules and reverse transcriptase assemble themselves in the host cell cytoplasm and then exit the host cell by forming vesicle structures from the host cell plasma membrane.
|
|
|
How is HIV transferred between people? What is the specific progression of events that normally occur in the human body after infection? What is AIDS and how do people normally die from this affliction?
|
HIV particles are transferred with the transfer of bodily fluids between humans. The most common means of transfer is unprotected sex but infection can also occur from sharing of unsterilized needles by intravenous drug users. Upon first infection, the HIV attacks a number of cells in the body provoking body defenses which include formation of antibodies against the HIV virus. The defenses of most infected individuals are able to fend off this initial infection and clear at least the blood stream of most viral particles. The remaining virus particles attack the T-cells of the immunoresponse system and over several years the T-cell count in the infected individual slowly declines. At some point the T-cell count becomes so low that individuals lose there ability to fend off many trivial infections and individuals develop acquired immunodeficiency syndrome (AIDS) and generally die for opportunistic infection by common infections such as pneumonia.
|
|
|
Compare the effect on the host cell of the lytic cycle versus the lysogenic cycle.
|
Lytic phages can only carry out lysis of the host cell, whereas lysogenic phages may either lyse the host cell or integrate into the host chromosome. In the latter case, the viral DNA (prophase is simply replicated along with the host chromosome. Under certain conditions, a prophage may exit the host chromosome and initiate a lytic cycle.
|
|
|
Bacterial reproduction is primarily by binary fission which a duplication division from a single cell. How does genetic diversity occur in bacterial cells that just undergo binary fission?
|
While binary fission is a replication process, genetic variability is introduced by mutation during DNA replication. Even though mutations are very rare there are so many bacteria and they divide so often that even the rarity of mutational events begins to add up. For example, there are so many E. coli cells in a human being that in total there would be 9 million mutation events per day in our human being.
|
|
|
What are the three mechanisms by which bacterial cells transfer DNA to other bacterial cells: Define and distinguish these three mechanisms from one another.
|
Transformation, transduction, and conjugation. In transformation, naked, foreign DNA from the environment is taken up by a bacterial cell. In transduction, phages carry bacterial genes from one bacterial cell to another. In conjugation, a bacterial cell directly transfers plasmid or chromosomal DNA to another cell via a mating bridge that temporarily connects the two cells.
|
|
|
For a bacterial cell, what is the advantage of regulating an enzyme pathway by gene expression over regulation of enzyme activity? What might be a disadvantage?
|
Regulating gene expression limits the waste of producing enzymes for a pathway that is not needed. Regulating gene expression requires more time to turn on a pathway than just regulation of enzyme activity. Synthesis of proteins takes more time than just activating or inactivating an enzyme that is already present in a cell.
|
|
|
What makes up the lac operon? How does the absence of lactose result in inhibition of the lac operon? How does the presence of lactose induce the lac operon?
|
An operon is a coordinately regulated cluster of genes with related functions. The lac operon consists of an operator gene and three genes for enzymes needed to metabolize lactose. A regulatory gene, separate from the lac operon genes, produces a repressor protein. In the absence of lactose, the repressor protein combines with an operator gene that is part of the lac operon. The operator gene is part of the promoter sequence of the operon upstream from the genes the produce the enzymes needed for metabolism of lactose. When the repressor protein combines with the operator, the transcription of the operon genes is blocked. In the presence of lactose, lactose molecules are converted to allolactose (an isomer of lactose) and the allolactose molecules bind to repressor protein molecules making them inactive. The repressor protein molecules are inactive in that they do not bind with the operator gene in the operon. Now RNA polymerase can bind with the operon promoter and transcribe the genes needed for lactose metabolism.
|
|
|
A mutation in E. coli changes the lac operator so that the active repressor cannot bind. How would this affect the cells production of β-galactosidase?
|
β-galactosidase is an enzyme that hydrolyzes lactose into its monmers, glucose and galactose and is one of the genes produced by the lac operon. If the repressor is unable to bind the operator gene, the cell would continuously produce β-galactosidase and the other two enzymes needed for lactose utilization even in the absence of lactose. This would be waste of cell resources.
|
|
|
What does the term "gene expression" mean in terms of a cell? What percentage of the genes in a human cell are expressed at any given time? What is the most important point of regulation of gene expression?
|
Gene expression means transcription and translation of specific genes. In most human cells, 5-20% of the total genes are expressed at any given time. The most important point of regulation of gene expression is the initiation of transcription.
|
|
|
What are the effects of histone acetylation and DNA methylation on gene expression? Once an mRNA reaches the cytoplasm of a eukaryotic cell, what are four mechanisms that can regulate the amount of active protein that can be produced from the sequence on the mRNA?
|
Histone acetylation is generally associated with gene expression while DNA methylation is generally associated with lack of expression. Degradation of the mRNA, regulation of translation, activation of the protein (by chemical modification for example) and protein degradation.
|
|
|
What is DNA cloning? What are the benefits of cloning genes into bacterial cells? What is a plasmid? What is the role of a plasmid in cloning?
|
DNA cloning is the insertion and expression of genes of interest into bacterial cells. Because bacterial cells can divide and grow rapidly, cloned cells can produce large amounts of copies of cloned genes as well as large amounts of their gene products. A plasmid is a small, circular piece of DNA found in many bacteria. The plasmid serves as a vehicle for inserting a gene or genes of interest into bacterial cells.
|
|
|
You want to study β-globin, an important protein found in human blood and to obtain sufficient amounts of the protein you clone the gene for β-globin. Describe the steps involved in cloning the gene. How will cloning the gene help you study the protein?
|
Identify and isolate the gene for β-globin from human blood cells. Isolate appropriate bacterial plasmids and cut and insert the β-globin gene into the plasmid. Reintroduce the the plasmids into bacterial cells making recombinant bacterial cells. Culture and harvest the recombinant bacteria. If manipulated correctly, the recombinant bacterial cells will produce large amounts of both the DNA for the β-globin gene and the β-globin protein which can be used for your studies.
|
|
|
What do we mean by the term "protein coding genes"? What percentage of the human genome is made up of protein coding genes?
|
Protein coding genes are DNA sequences that code for proteins (produce mRNA). Certain DNA sequences also code for rRNA and tRNA. Exons, DNA sequences that code for proteins, rRNA and tRNA make up about 1.5% of the human genome. One method of sequencing genome material is the "shotgun" method. Isolated copies of a given chromosome are degraded using restriction enzymes and the pieces of DNA are cloned producing separate bacterial colonies. The cloned DNA is isolated from the bacteria and then is sequenced. The sequences of all of the cloned fragments are then aligned using computers to produce the continuous sequence of the original chromosome.
|
|
|
How has DNA technology been used in the production of insulin and human growth hormone?
|
Deficiencies in insulin production can produce diabetes, an inability to properly regulate body glucose concentrations. Human growth hormone is required for normal development of humans. In both cases, affected individuals can live normally if given the deficient compound but natural sources are very limited and the compounds were expensive. The cloning of the genes for these molecules allows rapid and economical production of the proteins by bacteria. This has made insulin and human growth hormone plentiful and much more economical.
|
|
|
What is meant by the term "golden rice"? How can this product improve health, particularly in third world countries?
|
Golden rice is a strain of rice in which the genes for synthesis of beta carotene, a precursor for vitamin A have been cloned. The intention was to grow this rice in third world countries so that eating this rice would produce a natural supplement of vitamin A, particularly in regions where diets are deficient in Vitamin A.
|
|
|
In multicellular organisms, what are three examples of communication by direct contact?
|
Three examples of communication by direct contact are communication by transfer between animal cells through gap junctions, transfer between plant cells through plasmodesmata and cell-cell recognition.
|
|
|
In multicellular animals, what is the difference between paracrine and synaptic signals? Why are these types of signals considered to be local? What characterizes long distance signals in animal cells?
|
Paracrine signalling is an example of local communication that totally depends on chemical signals. Compounds are released from one cell, usually via the endomembrane system, and the compounds diffuse to adjacent cells where they are absorbed and stimulate reponses. Synaptic signals involve nerve cells and include an electrical component. Changes in membrane potential in a source cell causes the release of chemical signals that diffuse to an adjacent cell across a synaptic cleft where binding to the receptor membrane stimulates or inhibits changes in membrane potential. These signals involve transfer of signal molecules by diffusion from one cell to another. Long distance signals in animals involve the release of chemicals in one part of the body and transport through the bloodstream to receptor cells in another part of the body.
|
|
|
What are some of the general physiological characteristics of the “fight or flight” response in animals? What is a role of epinephrine in this response?
|
There is a) increased heart rate/blood pressure, b) constriction of blood vessels to non-essential organs - decreases flow, c) dilation of blood vessels in organs needed for exercise - increase flow, and d) blood glucose levels rise. Epinephrine is a hormone produced in the pancreas gland and released when stimulated by fear. The epinephrine moves in the bloodstream and binds with a number of cells including liver and muscle cells. The binding of epinephrine to liver and muscle cells stimulates the activity of glycogen phosphorylase which catalyzes the breakdown of glycogen to produce glucose. The glucose diffuses into the blood stream from liver cells that combined with the glucose released in muscle cells increases the substrate needed to stimulate respiration and the production of ATP to fuel physical activity.
|
|
|
Briefly describe what happens in the reception, transduction, and response stages of signaling at the cell level.
|
In reception, an external signal binds to a cellular receptor which may be located in the cell plasma membrane or cytoplasm. The binding causes a change in protein structure of the receptor molecule which in turn stimulates changes in other molecules. In transduction there is a cascade of changes in molecules stimulated by the original change in the cell receptor cell. This transduction pathway serves to amplify a signal event and provides a means for cellular regulation and coordination of response. The transduction pathway leads to changes in cell activity such as the increase in activity of certain enzymes and changes in protein synthesis: certain genes may be turned on and others turned off.
|
|
|
What kind of molecules are intracellular receptors? What chemical characteristics must a signal molecule possess to be able to bind with an intracellular receptor? Give an example of an intracellular receptor system.
|
Intracellular receptors are proteins. Signal molecules must easily diffuse across a cell membrane because they bind receptor proteins in the cytoplasm. The steroid hormone, testosterone is an example of a signal that binds to an intracellular receptor.
|
|
|
Give three examples of plasma membrane receptors. How are the names of these receptors related to their function?
|
Three examples of plasma membrane receptors are G-protein linked receptors, tyrosine kinase receptors, and ion channel receptors. The G-protein linked receptor protein stimulates a cytoplasmic G-protein, a protein whose ability to act as an internal signal depends on whether or not it is bound to a molecule of GTP. Tyrosine kinase receptors are membrane proteins that when bound to a signal molecule are converted to an active kinase or enzyme that stimulates phosphorylation of itself. Ion channels are gated membrane channels that open or close when they bind an external signal molecule.
|
|
|
What occurs during the Transduction phase of cell signaling? What is the function of a kinase and of a phosphorylase? What do kinases and phosphorylases have to do with signal transduction? A single molecule of epinephrine may bind with a liver cell. How is this signal amplified to produce large amounts of glucose?
|
Transduction is the cell response to a signal molecule binding to a receptor in either the cytoplasm or the plasma membrane. The complex formed by the signal and receptor triggers a cascade of molecular interactions that leads to metabolic or developmental responses in the cell. A kinase is an enzyme that generally phosphorylates (adds a phosphate) to a molecule and a phosphorylase is an enzyme that removes a phosphate from a molecule. Many signal transduction reactions involve phosphorylation and dephosphorylation of molecules. Each stage of a signal transduction pathway may trigger the production of multiple molecules for the next step in the pathway. For example in the pathway triggered by epinephrine, 100 molecules of active adenylyl cyclase can catalyse the production of 10,000 molecules of cyclic AMP. This amplification continues such that a million molecules of active glycogen phosphorylase catalyzes the formation of 100 million molecules of glucose-1-phosphate.
|
|
|
What are the three characteristics that define a natural compound as a hormone? What type of molecules can be hormones?
|
Hormone: a) The compound is synthesized in one part of the organism and transported to cells that are the site of activity. b) The hormone molecule interacts with a specific plasma membrane receptors. c) The molecule stimulates cellular activity at very low concentrations (micromolar or less. Hormones include : proteins, small peptides, amino acid derivatives and steroids.
|
|
|
What is phototropism in plants? What plant hormone mediates phototropism? Specifically, how is the phototropic response caused by a plant hormone?
|
Phototropism is a growth response where a plant stem grows towards a directional light source. Phototropism is mediated by the plant hormone auxin. Auxin is produced in the apical meristem and the directional light source shines on one side of the growing stem stimulating the transport of auxin from the lighted side to the dark side. In the proper concentration, auxin stimulates cell elongation so that cells on the dark side of the stem elongate more that cells on the lighted side of the stem. This differential expansion causes the stem to bend toward the light source.
|
|
|
What is apical dominance? What are thought to be the roles of auxins and cytokinins in controlling apical dominance.
|
In apical dominance, growth is greatest at the terminal bud of a stem and lateral bud growth is inhibited (producing very little branching). This causes the plant to have the appearance of a pine tree (an example of apical dominance). Auxin produced at the apical meristem diffuses down the stem and inhibits the development of a lateral buds and therefore branching. It is thought that the auxin inhibits the production of cytokinins in the lateral buds. Cytokinins are thought to stimulate cell division in lateral buds and therefore growth of branches.
|
|
|
What is the “foolish rice seedling” disease and how did its discovery relate to gibberellins? How do Mendel's tall and dwarf pea varieties differ in their production of gibberellin and why?
|
"Foolish rice seedling" disease is rapid and abnormal growth of rice plants producing long weak stems. These stems often collapse onto the ground leading to damage of the stem and any seed it produces. This disease was correlated with rice infection by a fungus which produces high concentrations of the hormone gibberellin. After gibberellin was identified in the fungus it was also found to naturally occur in plants and in the right concentration it naturally regulates normal stem elongation. Mendel's tall pea plants produce normal concentrations of gibberellin and normal growth of pea stems. Dwarf pea plants have a mutation in a gene for an enzyme required in the biosynthetic pathway for gibberellin. Dwarf plants have very low concentrations of gibberellin precursors and therefore there is very little growth of their stems.
|
|
|
How are drought stress and abscisic acid concentration thought to be related in plants? How does abscisic acid affect stomatal opening? Why would this be advantageous under drought conditions?
|
Abscisic acid concentration (ABA) increases in plants under drought stress. Increasing ABA concentration closes stomates. Stomates regulate water loss through the leaf epidermis and under drought conditions closing stomates would allow plants to better conserve water.
|
|
|
What plant hormone is related to the old saying "one bad apples spoils the lot"? Why? What is the experiment with tomato that supports the role of this plant hormone in fruit ripening?
|
Ethylene stimulates fruit ripening and ethylene synthesis. If an apple stored with other apples begins to ripen (becoming soft and mushy) it will synthesize and release ethylene. Since ethylene is a gas it will diffuse to the other apples and stimulate ripening and ethylene synthesis in them, accelerating the ripening process. A definitive experiment with tomatoes was the production of antisense mutants for an enzyme necessary for ethylene synthesis. These mutants could not synthesize ethylene and their fruit remained green and would not ripen. Putting the unripened fruit of these plants in a container with ethylene stimulated normal ripening.
|
|
|
Where in the body are the following endocrine glands: hypothalmus, posterior pituitary, thyroid, parathyroid, and pancreas? Where are ADH, calcitonin, parathryoid hormone (PTH), insulin, and glucogon produced? What kind of molecules are these hormones?
|
The hypothalmus and posterior pituitary are found at the base of the brain. The thyroid and parathyroid glands are found in the throat. The parathyroid glands are on the thyroid glands. The pancreas is found between the kidneys. ADH (a peptide) is synthesized in the hypothalmus and stored in the posterior pituitary gland. Calcitonin (a peptide) is synthesized in the thyroid gland and PTH (a peptide) is synthesized in the parathyroid gland. Both insulin and glucagon are synthesized in the pancreas.
|
|
|
In a desert with little to drink, a man becomes dehydrated. How does this change his blood osmolarity? Explain what would happen in the man’s body that would maintain homeostasis of blood osmolarity. How could the man end up with a blood osmolarity that was too low? How would the body respond to return the osmolarity to homeostasis in this case?
|
Dehydration normally leads to a concentrating of solutes in the blood or an increase in blood osmolarity. The increase in osmolarity is detected in the brain and the hypothalmus directs the pituitary gland to release antidiuretic hormone (ADH) into the blood stream. ADH causes an increase in water permeability in cells in the kidneys and water is reabsorbed and returned to the blood system. The brain also signals the body that it is thirsty and this will result in drinking water which should also result in dilution of blood concentration. Low osmolality or diluted blood concentration is most often the result of drinking too much water. In this case the hypothalmus directs the pituitary gland to lower the release of ADH which leads to a decrease in water permeability in the kidneys, more water is lost as urine and the concentration of solutes increases back to the acceptable level. The brain also signals the body that its thirst is quenched which should lead to a reduction in drinking of liquids.
|
|
|
Give three steps in a negative feedback loop. Describe how a house air conditioner illustrates the concept of a negative feedback loop.
|
There is a trend in change of the value of some process. When this trend leads to a change of sufficient magnitude, the change is detected by a sensor. The sensor then directs an effecter to counter the change and the process value returns to its original value. A house includes a thermometer which is the sensor and a compressor that produces cold air which is the effecter. The temperature is set by the thermostat. In the summer the warm outside air causes the house air to warm up and the increase in temperature is the trend away from the original set value. When the temperature heats up to some maximum, the thermostat directs the compressor, which is the effecter, to turn on and produce cold air. The cold air counters the trend of warming air and the air temperature is reduced until it is back to an acceptable value.
|
|
|
What are some of the important functions of calcium in the human body? Calcium concentration in the blood system is maintained at about 1 mM. What are the two hormones that regulate blood calcium concentration? How do these two hormones regulate calcium concentration by negative feedback loops?
|
Calcium serves three major functions: 1) it is a significant component of bones, 2) it serves as a cell signal and 3) it is a cofactor for a number of proteins and enzymes. As a cell signal it directs constriction and relaxation of blood vessels, transmission of nerve impulses, and contraction of muscles. The two hormones that regulate blood calcium concentration are calcitonin and PTH (parathyroid hormone). When calcium concentrations become to high in the blood, calcitonin is released by the thyroid gland. Calcitonin causes bone cells to increase the rate of calcium deposition into bones. The intestines and kidney cells are signaled to decrease the rate of reabsorption of calcium. and therefore there is an increase in calcium excretion in urine and a decrease in blood calcium concentration back to homeostasis value. If calcium concentrations drop too low, PTH is released by the parathyroid gland. PTH signals the opposite responses: Bones are signaled to breakdown calcium and release it into the blood and cells in the kidneys and intestines are signaled to increase the rate of absorption of calcium and this leads to an increase in blood calcium concentration.
|
|
|
Why is glucose important to animals? Describe the two feedback loops that maintain a relatively constant glucose concentration in the blood. Which of the feedback loops would be active in someone who just ate a big meal, with lots of sugars? Which of the feedback loops would be active in someone who had not eaten in twelve hours?
|
Glucose is the major carbohydrate input into respiration production in animal cells. Glucose is oxidized and the energy released in this process is trapped as ATP which is needed for running most processes in cells. Glucose also serves as a source of carbon backbones for the synthesis of different types of molecules including proteins and lipids and phospholipids. The two negative feedback loops that control blood glucose concentration are insulin and glucagon. Insulin is produced in certain cells in the pancreas and insulin is released into the blood stream when calcium concentration becomes too high. Insulin stimulates cells to take up glucose from the blood stream and liver cells in particular to take up glucose and make the storage carbohydrate glycogen. This reduces the glucose concentration in the blood back to acceptable values. Glucagon is also produced by certain cells in the pancreas and is released into the bloodstream when glucose concentrations become too low. Glucagon has the opposite effect as insulin: liver cells are stimulated to breakdown glycogen and release glucose into the blood causing an increase in blood glucose back to homeostasis values. Someone who ate a big meal should release insulin and stimulate the uptake of glucose into the blood system this would also reduce the release of glucagon. Someone who had not eaten would have increasing concentrations of glucagon stimulating the release of glucose into the blood as well as decreasing concentrations of insulin.
|
|
|
What is diabetes and what is the difference between type I and type II diabetes?
|
Diabetes is the name of the condition where the body allows the concentration of glucose to become very high in the blood stream. This is potentially very dangerous for a number of reasons including producing malfunction of the kidneys which are forced to remove the excess glucose from the blood stream. In Type I diabetes, there is an autoimmune response where the body's immune system attacks and destroys the pancreas cells that produce insulin. Therefore individuals with Type I are deficient in insulin. Type II diabetes is most commonly due to a change in receptor cell sensitivity to insulin and therefore these individuals have enough insulin, it just does not elicit the normal response. There are also individuals with Type II diabetes that do not have enough insulin and this is not generally due to an autoimmune response.
|
|
|
The broad region that corresponds to the length of the thick filaments of myofibrils.
|
A band
|
|
|
A type of muscle that forms the contractile wall of the heart. Its cells are joined by intercalated disck that relay each heartbeat.
|
cardiac muscle
|
|
|
The area near the edge of the sarcomere where there are only thin filaments.
|
I band
|
|
|
A single motor neuron and all the muscle fibers it controls.
|
motor unit
|
|
|
A fibril collectively arranged in longitudinal bundles in muscle cells (fibers); composed of thin filaments of actin and a regulatory protein and thick filaments of myosin.
|
myofibril
|
|
|
The thick and thin filaments that form the myofibrils.
|
myofilaments
|
|
|
An oxygen-storing, pigmented protein in muscle cells.
|
myoglobin
|
|
|
The process of progressively increasing the tension of a muscle by activating more and more of the motor neurons controlling the muscle.
|
recruitment
|
|
|
The fundamental, repeating unit of striated muscle, delimited by the Z lines.
|
sarcomere
|
|
|
A specialized endoplasmic reticulum that regulates the calcium concentration in the cytosol.
|
sarcoplasmic reticulum (SR)
|
|
|
The theory explaining how muscle contracts, based on change within a sarcomere, the basic unit of muscle organization, stating that thin (actin) filaments slide across thick (myosin) filaments, shortening the sarcomere. The shortening of all sarcomeres in a myofibril shortens the entire myofibril.
|
sliding-filament model
|
|
|
A type of muscle lacking the striations of skeletal and cardiac muscle because of the uniform distribution of myosin filaments in the cell.
|
smooth muscle
|
|
|
The maximal, sustained contraction of a skeletal muscle, caused by a very fast frequency of action potentials elicited by continual stimulation.
|
tetanus
|
|
|
A filament composed of staggered arrays of myosin molecules; a component of myofibrils in muscle fibers.
|
thick filament
|
|
|
The smaller of the two myofilaments consisting of two strands of actin and two strands of regulatory protein coiled around one another.
|
thin filament
|
|
|
Infoldings of the plasma membrane of skeletal muscle cells.
|
transverse (T) tubules
|
|
|
The borders of a sarcomere.
|
Z lines
|
|
|
What are the major parts of a neuron? Functionally, what is the difference between an axon and a dendrite? In a vertebrate neuron, what is the axon hillock? What is the function of astroctyes, and radial glia? Where are these cells found? Where do you find the myelin sheath and how is the sheath related to Schwann cells?
|
The major parts of a neuron are the cell body (which contains the nucleus and other organelles), the processes and the synapse. Dendrites and axons are the processes and dendrites transmit signals away from their tips towards the cell body while axons transmit signals away from the cell body towards their tips. The axon hillock is location on the axon that is closest to the cell body. Astrocytes and radial glia, cells found in the central nervous system, are support cells that support neurons structurally and functionally. The myelin sheath surrounds the axon of certain vertebrate neurons. Individual Schwann cells surround the axon at discrete intervals producing the myelin sheath.
|
|
|
What is meant by the term resting potential in terms of a neuron? What would be a common value for the resting potential of a neuron? What are the relative concentrations of Na+ and K+ across the membrane in an unstimulated neuron (at resting potential)? How does an increase in Na+ diffusion affect the resting potential? How does an increase in K+ diffusion affect the resting potential? What is the role of the sodium-potassium pump in maintaining the resting potential of a neuron?
|
The resting potential of a neuron is the membrane potential when the neuron is not being stimulated. Commonly, the resting potential of neurons is between -60 and -80 mV, indicating the inside of the cell is negative relative to the solution outside the cell. At resting potential there is a high concentration of sodium ions outside the cell and a high concentration of potassium ions inside the cell. Typical values for sodium ions would be 150 mM outside the cell and 15 mM inside while the concentrations for potassium ions would be 150 mM inside and 5 mM outside. An increase in Na+ diffusion would make the inside more positive and the membrane potential would move towards zero. An increase in K+ diffusion would make the inside more negative and the membrane potential would become a more negative value. Sodium and potassium ions diffuse across the membrane in response to their respective concentration gradients. The sodium-potassium pump requires energy and moves the sodium and potassium ions against their concentration gradients to maintain the gradients described above.
|
|
|
Why are neurons termed "excitable" cells? What do hyperpolarization and depolarization mean in terms of the neural membrane?
|
Neurons are termed "excitable" cells because stimulation can induce large changes in membrane potential. In hyperpolarization the membrane potential becomes more negative and in depolarization the membrane potential moves towards zero (becomes more positive).
|
|
|
Define graded potential, threshold potential and action potential. How do graded potentials and action potentials differ? For graded potentials, what ions cause hyperpolarization, what ions cause depolarization? Why?
|
Graded potential - change in potential the does not cross the threshold potential and for which the magnitude is proportional to the strength of the stimulus. Threshold potential - a membrane potential at which potential changes switch from graded to action potential. Action potential - an all or nothing change in potential. The magnitude of an action potential is independent of strength of a stimulus.
|
|
|
What is a voltage-gated channel? At the resting potential of a membrane, what is the condition of voltage-gated channels for Na+ and Cl-?
|
A voltage-gated channel is an ion channel that can be either open or closed and the opening or closing is regulated by the voltage of the membrane. At the resting potential, both Na+ and K+ voltage-gated channels are closed.
|
|
|
How are signals propagated along a neuron? Why are action potentials only triggered in one direction? How are signals propagated so rapidly along a myelinated vertebrate neuron?
|
The triggering of an action potential in one location triggers an action potential in the neighboring area towards the synaptic cleft (along the axon). Potentials do not go backwords because the membrane location where an action potential has just occurred cannot re-trigger while it is repolarizing. Myelinated axons of vertebrate neurons possess open spaces (not myelinated) at regular intervals. Action potentials are triggered in an open space and ions diffuse on the inside of the axon to the next open space where another action potential is triggered. This is faster than if the axon was bare and each action potential had to trigger an adjacent action potential.
|
|
|
Define the terms synaptic cleft and neurotransmitter.
|
A synaptic cleft is the space between the membrane of a presynaptic axon tip and a postsynaptic neuron membrane. A neurotransmitter is a chemical signal that is released by the presynaptic membrane of a stimulated neuron and diffuses across the synaptic cleft where it binds to and stimulates the postsynaptic cell.
|
|
|
In a chemical synapse, how do action potentials in a presynaptic cell effect Ca+2 concentration and the release of a neurotransmitter into a synaptic cleft? What are ligand-gated ion channels? What is their role in transmission of signals from a presynaptic cell to a postsynaptic cell?
|
The action potential depolarizes the membrane of an axon opening calcium channels which results in an increase in cytoplasmic calcium concentration. The elevated calcium concentration stimulates the fusion of vesicles holding neurotransmitter molecules with the plasma membrane. The neurotransmitter is released to the outside of the cell in the synaptic cleft. Ligand-gated ion channels are ion channels that open when they bind some type of signal molecule. In many cases neurotransmitter molecules serve as the signal or ligand that binds ligand-gated ion channels involved in neuron response. Thus, ligand-gated ion channels in the postsynaptic neuron membrane bind with neurotransmitter molecules released by the presynaptic neuron. Binding causes the ion channels to open and ion transport through the channel alters the ion concentration inside the postsynaptic cell which can lead to depolarization and triggering of an action potential.
|
|
|
Arrange the following terms in order from the most general and encompassing level of organization to the most specific level: bundle of fibers, cell, muscle, myofibril, sarcomere. In the sarcomere, what is the role of actin and myosin in muscle contraction?
|
Muscles are made up of a bundle of muscle fibers with each fiber being a muscle cell. In each cell there are multiple myofibrils which are bundles of actin and myosin proteins. The actin and myosin proteins are arranged in sequential units called sarcomeres which are capable of shortening and producing muscle contraction. Muscle contraction is produced when actin filaments (thin filaments) are pulled together by myosin filaments (thick filaments) Muscle contraction occurs when myosin (thick) filaments pull actin (thin) filaments and shorten the length of a sarcomere. Myosin is a motor protein and with the input of ATP, it attaches to actin filaments on each of its ends and basically pulls the overlapping actin filaments.
|
|
|
Motor neurons cause muscle contraction in muscle fibers by producing the release of calcium from the sarcoplasmic reticulum. Starting with an action potential in a neural axon, what are the steps that lead to the release of the calcium ions? How does calcium lead to muscle contraction? What causes muscle relaxation to occur?
|
The action potential stimulates the release of the neurotransmitter, acetylcholine, which diffuses across the synaptic cleft to the muscle fiber cell. The acetylcholine binds with receptor proteins on the muscle cell which triggers an action potential. The action potential is propagated along the plasma membrane of the muscle cell and down the membrane-lined T-tubule into the cytoplasm through the sarcoplasmic reticulum which is a calcium reservoir surrounding a myofibril. The action potential along the T-tubule changes the membrane permeability of the sarcoplasmic reticulum which releases calcium into the cytoplasm of the muscle fiber. Without calcium, the protein tropomyosin covers myosin binding sites on actin molecules in the myofibril. This blocks interactions between myosin and actin. When calcium concentration increases in the cytoplasm, calcium binds to the tropin binding complex on the tropomyosin protein causing the tropmyosin protein to change conformation and expose the myosin binding sites on the actin molecules. With these sites exposed and the presence of ATP, myosin binds to the actin and slides the actin filaments together in myofibrils causing muscle contraction. Pumps in the sarcoplasmic reticulum lower the calcium concentration around the actin and myosin proteins until calcium is removed from the tropin binding complex. Removal of the calcium causes the tropomyosin protein to change conformation and again cover the myosin binding sites and the myosin is released from the actin, releasing the muscle contraction.
|
|
|
Graph an action potential as a plot of membrane potential (y-axis) vs. time (msec). Identify threshold potential, and the following phases of an action potential: depolarization, rising, falling, and undershoot on your graph. Describe what is happening in the axon membrane at each of these phases in terms of ion channels and ion movement.
|
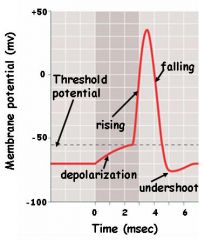
Depolarization - stimulus causes a graded rise in membrane potential as Na+ channels are opened. When the threshold is reached, the opening of all gated Na+ channels stimulates an action potential. Rising phase is marked by influx of enough Na+ ions that the inside becomes positive relative to the outside. Falling phase begins when membrane potential reaches the point that closes inactivation gates of Na+ channels and opens activation gates of most K+ channels. K+ ions leaving the cell causes the inside to become more negative and the potential to fall. Undershoot phase - there is a continuation of the drop in membrane potential because some K+ gates are still closing. K+ channel gates close, inactivation gates on Na+ channels open and membrane potential returns to resting state.
|

