![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
25 Cards in this Set
- Front
- Back
|
The most common ways of making cells competent are:
|
• Chemical treatment
• Heat shock • Electroporation • Biolistics • Sonication |
|
|
competent cells:
|
Cells that can take up DNA from their environment
|
|
|
Transformation
|
Competent cells take up extracellular DNA through this process.
(or transfection if a viral vector is being used) |
|
|
Methods to introduce genes into ALL the cells of a multicellular animal
|
Microinjection
Nuclear transplantation |
|
|
Nuclear transplant
|
In this process, the whole nucleus (with all of its genetic information) is swapped or “switched out”
Eggs (NOT embryos) are fertilized with transgenic sperm nuclei by injecting the nuclei into the eggs This is also a tricky process because many eggs get more than one sperm cell (leading to cell death) |
|
|
Microinjection
|
DNA is injected directly into the nucleus of a fertilized egg or a very early embryo using a VERY fine pipette
After microinjection, DNA is naturally integrated into the chromosome creating a transgenic animal The transformed fertilized egg is implanted into an animal for the completion of development This is a tricky process with limited results |
|
|
Organisms frequently used in whole organism genomic experiments
|
• Mice are commonly used for laboratory experiments, but they are expensive and microinjection is tricky
• Frogs are beginning to be more widely used because they are cheap, have 100s of eggs, and develop quickly ex utero |
|
|
Selectable markers
|
Antibiotic resistance markers: most common
Only cells with the recombinant DNA can grow on/in the presence of the antibiotic Growth on an antibiotic medium only selects for the plasmid (successful transformation, but not insertion of our gene of interest) |
|
|
α-complementation
|
If the MCS is located within a gene (lacZ), then successful insertion of YFG will disrupt lacZ expression.
Allows us to select for sucessful plasmid transfection and YGF insertion at the same time |
|
|
What does lacZ do?
|
When grown in the presence of X-gal:
colonies expressing lacZ (YFG absent): cleave X-gal, colonies are blue! colonies not expressing lacZ (YFG present): no X-gal cleavage, colonies are white We will select white colonies |
|
|
Screening
|
o This is the final step of the cloning process
• It is extremely broad because each type of screen is specific for your area of interest • This process equates to finding the needle in the haystack |
|
|
constructing a cDNA library
|
• is constructed from mature mRNA so it has only coding information
• Isolate total cellular RNA • Hybridize RNA fragments with poly-T oligos (oligo (dT)) • Wash to discard cellular debris (100mM NaCl) • Add buffer to reverse poly-T oligo binding and elute mRNA • Hybridize with oligo(dT) primer with RE recognition site • Add reverse transcriptase (enzyme from retroviruses; make DNA from RNA), and methylated bases • Add RNase H (nicks in RNA stand to fool DNA Polimerase into thinking there are a lot of primers) • Add DNA polymerase and dNTPs to synthesize second DNA strand • Cut cDNA with restriction enzymes and mass ligate all cDNAs into plasmid vectors |
|
|
Using a cDNA library
|
This collection of fragments is used to isolate specific genes and other DNAs
• A gene is a book and screening is searching for that book in a library using only a fragment of a sentence o The fragment of a sentence is your probe o It is possible to screen a library by determining if your probe will hybridize to a sequence within the library o If the probe hybridizes to the blot, you have found the gene that corresponds to the DNA sequence |
|
|
Hybridization: what is it, and who does it?
|
• Hybridization is simply allowing two things to be drawn together by their natural affinity for one another
o Complementary DNA strands hybridize o Complementary DNA and RNA strands hybridize o Proteins and DNA can hybridize o Antibodies and proteins can hybridize |
|
|
oligo(dT)
|
poly T oligos that grap onto the polyA tails of mRNA.
|
|
|
Southern Blot steps
|
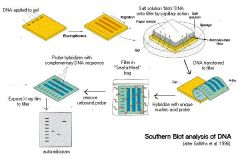
o All genomic DNA fragments are separated by size
o DNA smears are transferred to a membrane o Incubate blot with radioactively labeled probe to hybridize complementary sequences |
|
|
Southern blot
|
• In a Southern blot, genomic DNA is hybridized with probe DNA to see if DNA-DNA hybridization occurs
• Genomic DNA is digested with REs and separated by agarose gel electrophoresis • The DNA smear is transferred to a membrane (“blotted) and then incubated with a probe to detect certain sequences • Southern blotting is used to detect the presence of sequences within the DNA or to “fingerprint” individuals |
|
|
Northern blotting
|
• a similar technique to Southern blotting used for detecting RNA
• Usually used to measure gene expression levels by detecting mRNA transcripts |
|
|
Microarray
|
• A technologically advanced version of the reverse experiment is called a microarray
• Green is on, red is off • Measuring the expression of genes on a microchip allows for a tremendous number of genes to be analyzed at once |
|
|
Example: We saw that a certain protein was found in elevated levels in disease tissue; need gene for study
|
Got a little bit of DNA sequence from the protein’s amino acids (to use as a probe later)
Constructed a cDNA library from various tissues Probed genomic DNA from each diseased tissue type using Southern blot analysis Screened only the cDNA libraries with our gene of interest for clones containing sequence from that gene Isolated those clones Now we must sequence the DNA to get the mutated gene seqence; Why can’t we just BLASTn for sequence? • Because it is mutated |
|

Automated fluorescent dideoxy DNA sequencing
|

Sequencing is now an automated process using capillary tubes, fluorescent nucleotides, and laser detection methods
As the labeled DNA fragments elute off the capillary gel, they are pulsed by a scanning laser |
|
|
Sanger/ Dideoxy DNA Sequencing
|
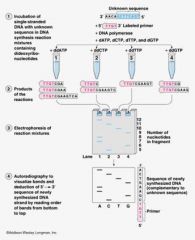
• Start with dsDNA
• Make ssDNA template • Set up 4 reactions, one tube for each base o Add nucleotide mix with dideoxy bases, which stop DNA compliment formation at that base. Ex: ddTTP stops stand at A. |
|
|
PCR
|
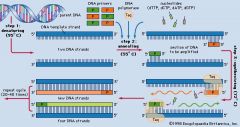
If we want to make a great deal of DNA, or modify it in many ways, we can utilize the polymerase chain reaction
Flanking primers amplify DNA sequence Because the sample is heated, a thermo- stable DNA polymerase is necessary |
|
|
western blot steps
|
1) isolate total protein
2) separate by size in an DSD- polyacrilimide gel 3) transfer separated proteins to nitrocellulose membrane 4) block membrane 5) primary antibody binds to our protein 6) secondary antibody binds to primary antibody o contains chemiluminescent conjugate 7) develop 8) visualize and quantify with digital |
|
|
Reporter genes
|
Reporter genes produce detectable traits while the cell is still alive and well; much more informative
Green fluorescent protein (GFP) is one of the most widely used reporter genes |

